HLA molecular mismatch analysis in patients receiving kidney and the hematopoietic stem / facilitating cell induction protocol
Anat Tambur1, Cynthia Garcia-Sanchez1, Crystal Y. Usenko1, Vasilis Kosmoliaptsis2, Hannah Copley2, Joseph R. Leventhal1, Suzan Ildstad3.
1Northwestern University, Chicago, IL, United States; 2Surgery, Cambridge, Cambridge, United Kingdom; 3U of Louisville, Louisville, KY, United States
Introduction: The purpose of this work was is to evaluate a potential role for HLA molecular mismatch (MM) analysis in predicting outcomes in a cohort of HLA-mismatched, related and unrelated recipients of combined living donor kidney and hematopoietic stem cell transplants (HSCT), performed to establish transplantation tolerance.
Cohort: Recipients underwent nonmyeloblative conditioning followed by a manipulated HSCT enriched for facilitating cells (FCR001) as previously described (Leventhal et al STM 2012). 32/37 donor/recipient pairs enrolled in this study met the inclusion criteria for our analysis (availability of unambiguous high resolution HLA typing).
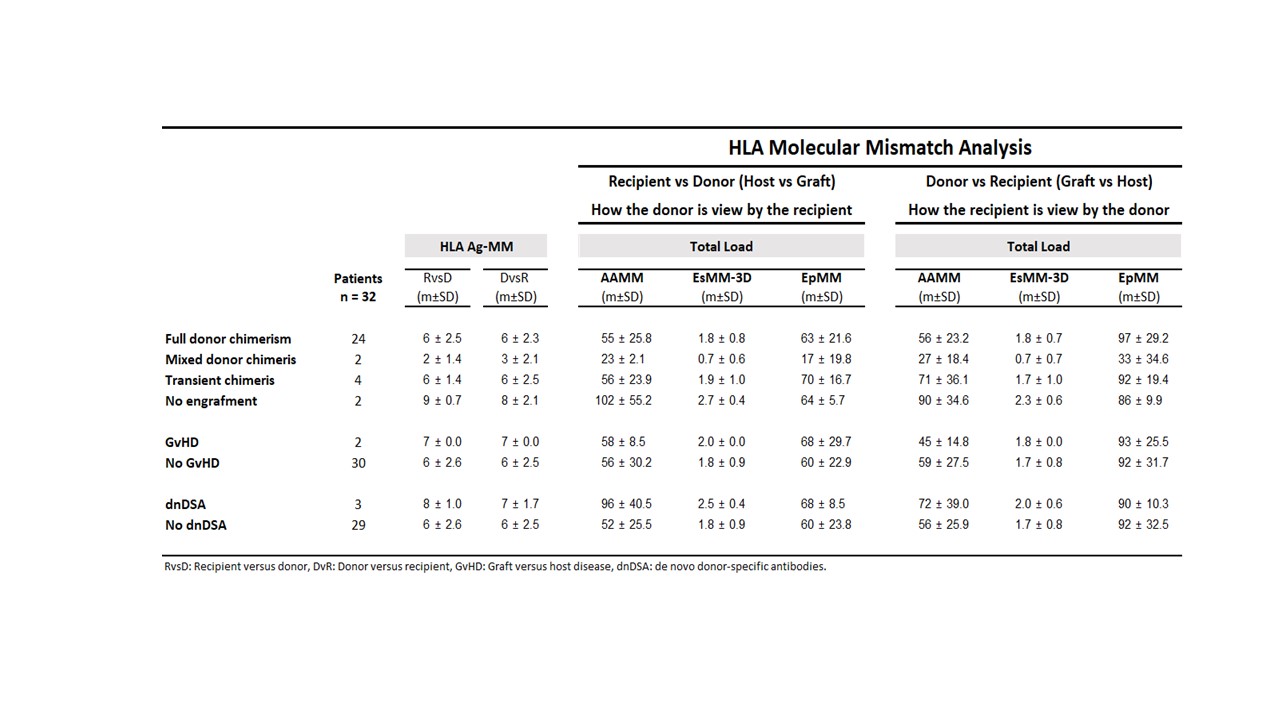
Analysis: HLA-A,-B,-C,-DRB1,-DQA1,-DQB1 MM load was determined using 3 approaches: amino-acid count (AAMM); electrostatic MM (ESM-3D) and HLA-Matchmaker/Eplets (EpMM). Outcome parameters included level of stem cell engraftment (chimerism); kidney rejection; dnDSA formation and graft versus host disease (GvHD). Analysis was performed for both host vs graft (HvG) and graft vs host (GvH) directions.
Results: Average age was 42 ± 11 y, 25 patients were male. 24/32 patients achieved full durable donor chimerism (>98% by STR analysis), 2 persistent mixedchimerism, 4 transient chimerism and, only 2 patients had lack of stem cell engraftment. GvHD occurred in 2 patients; 3 patients developed dnDSA, and 4 patients had biopsy proven kidney rejection.
The 2 patients that did not engraft showed higher AAMM and ESMM-3D scores compared with patients with durable engraftment (102± 55vs 55 ± 26 and 2.7 ± 0.4 vs 1.8 ± 0.8, respectively) though their EpMM was virtually the same (64 ± 5.7 vs 63 ± 21). 2 patients with full donor chimerism developed GvHD. Both had 7/10 HLA antigen MM in the GvH direction compared with 6 ± 2.6/10 MM for no GvHD patients; one of them died. Mean AAMM, ESMM-3D, and EpMM in either GvH or HvG directions were similar between these patients and the rest of the cohort. 3 patients developed dnDSA, HvG direction analysis showed higher AAMM, ESM-3D, and EpMM compared with no dnDSA patients (96 ± 41, 2.5 ± 0.4 and 68 ± 9 vs 52 ± 26, 1.8 ± 0.9, and 60 ± 24, respectively).
Conclusion: It appears that Molecular Mismatch strategies, currently used to predict outcomes after SOC kidney transplant, are not useful in predicting outcomes in combined kidney + FCR001 patients with regards to engraftment or GvHD risk. A previously reported correlation between dnDSA and increased mismatch load, was confirmed. Other approaches to assess MM at the indirect presentation level should be explored. Importantly, new tools to assess immunogenicity beyond number/load of mismatch should be developed.