
DEEPGRAFT, a deep learning algorithm to classify kidney transplant diseases from digital whole slide biopsy images
Jesper Kers1,2,3, Roman Bulow4, Barbara Klinkhammer4, Gerben Breimer1, Rianne Hofstraat1,3, Garry Corthals3, Hessel Peters-Sengers5, Tobias Pieters6, Frederike Bemelman7, Joris Roelofs1,2, Sandrine Florquin1,2, Tri Nguyen8, Jakob Kather9,10, Peter Boor4.
1Pathology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands; 2Pathology, Leiden University Medical Center (LUMC), Leiden, Netherlands; 3Van ‘t Hoff Institute for Molecular Sciences (HIMS), University of Amsterdam, Amsterdam, Netherlands; 4Institute of Pathology, University Hospital RWTH Aachen, Aachen, Germany; 5Center for Experimental & Molecular Medicine (CEMM), Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands; 6Department of Nephrology, University Medical Center Utrecht, Utrecht, Netherlands; 7Renal Transplant Unit, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands; 8Department of Pathology, University Medical Center Utrecht, Utrecht, Netherlands; 9Department of Medicine III, University Hospital RWTH Aachen, Aachen, Germany; 10German Cancer Consortium (DKTK), German Cancer Research Center (DFKZ), Heidelberg, Germany
DEEPGRAFT consortium.
Introduction: The gold standard and currently only means to specifically classify diseases of renal allografts is a histopathological assessment of a renal transplant biopsies according to the latest Banff criteria. Some disagreements, however, exist among the transplant pathologists on certain lesions and even diagnoses, which might potentially result in differences in treatment and subsequent graft outcome. Deep learning using convolutional neural networks (CNNs), is a highly effective computational approach, able to recognize patterns in histopathological whole slide images (WSIs) with the potential of automated and highly reproducible classification of post-transplant diseases. The aim of the DEEPGRAFT study is to develop, deploy and validate CNNs to improve renal transplant biopsy-based disease classification and prognostication.
Materials and Methods: DEEPGRAFT is an international multicenter consortium that includes the protocol and for cause biopsies from three centers in Europe (Amsterdam UMC, UMC Utrecht and RWTH Aachen). For each biopsy, the H&E, Silver and PAS staining was digitized (N = 3159 WSIs in total). Cases were labeled to the following classes according to the pathology reports: 1) T cell-mediated rejection; TCMR, 2) antibody-mediated rejection; ABMR, 3) mixed rejection, 4) viral nephropathy, 5) no specific allograft pathology, and 6) other diseases of the allograft. A shufflenet was trained by transfer learning and was validated by patient-level three-fold cross-validation. Area-under-the receiver operating characteristics (AUROC) was used as the primary endpoint. We trained a model with all three stains and compared it to a model trained on only the PAS staining as input. The complete tissue (cortex and medulla) was used as input.
Results: The CNN trained on the PAS stains had the highest discrimination index for stable graft histology (AUROC = 0.83), which improved when all three stains were included (AUROC = 0.88), see Figure.
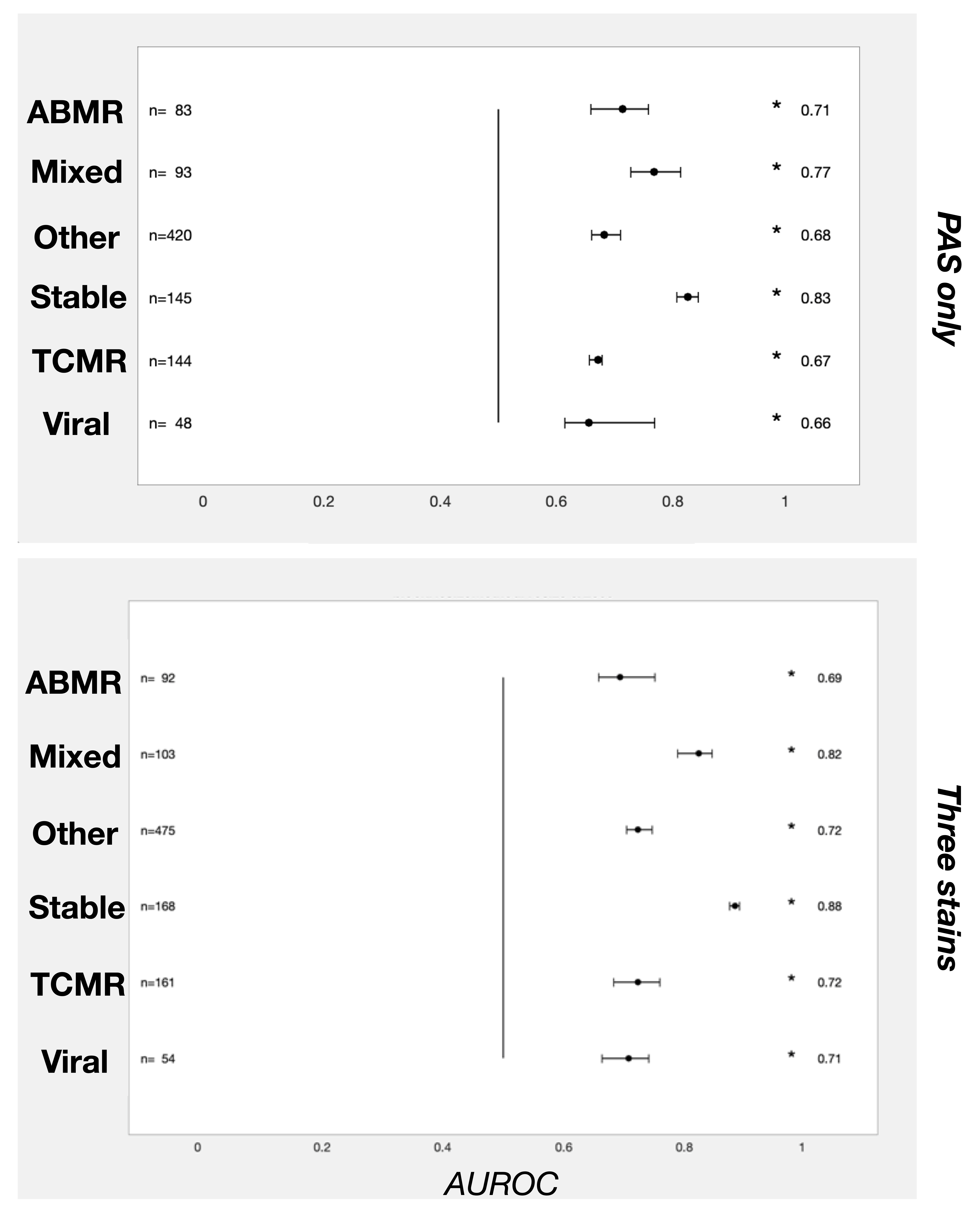
Discrimination indices for TCMR, ABMR and mixed rejection ranged from 0.67 – 0.77, respectively, and improved to 0.69 – 0.82. Noteworthy, the CNN was able to discriminate viral nephropathies from the rejection classes (AUROC = 0.71), even though no additional immunohistochemistry was used as input for the networks.
Discussion: We have shown the potential of a deep learning algorithm to discriminate between clinically relevant diagnoses only from histopathological images. The integration of several histological stainings improved the performance. Our study provides the first example of a large multicenter trial examining the potential of artificial intelligence on non-tumor pathology. Next, we will further extend and refine the algorithm in even larger cohorts and using additional data integration.
Conclusion: Deep convolutional neural networks have the potential to reproducibly classify transplant histopathology, even without human expert annotation of individual lesions.
Dutch Kidney Foundation Kolff Junior Postdoc grant, 17OKG23. University of Amsterdam, Human(e) AI Research Priority Area Seed grant.
