Novel human kidney cell subsets identified by Mux-Seq
Andrew W. Schroeder1, Swastika Sur1, Priyanka Rashmi1, Izabella Damm1, Arya Zarinsefat1, Matthias Kretzler4, Jeff Hodgin4, Tara Sigdel1, George Hartoularos2, Jimmie Chen Ye2, Minnie M. Sarwal1,3.
1Surgery, University of California San Francisco, San Francisco, CA, United States; 2Institute of Human Genetics, University of California San Francisco, San Francisco, CA, United States; 3Medicine, University of California San Francisco, San Francisco, CA, United States; 4Medicine, University of Michigan, Ann Arbor, MI, United States
Kidney Precision Medicine Project.
Introduction: The kidney is a highly complex organ that performs multiple functions necessary to maintain systemic homeostasis, with complex interplay from different kidney sub-structures and the coordinated response of diverse cell types, few known and likely many others, as yet undiscovered. Traditional global sequencing techniques are limited in their ability to identify unique and functionally diverse cell types in complex tissues.
Materials and Methods: Herein we characterize over 45,000 cells from 12 human kidneys including normal native as well as transplant kidney biopsies using unbiased single-cell RNA sequencing. We also apply, for the first time, an approach of multiplexing kidney samples (Mux-Seq), pooled from different individuals, to save input sample amount and cost. We applied the computational tool Demuxlet to assess differential expression across multiple individuals by pooling human kidney cells for scRNA sequencing, utilizing individual genetic variability to determine the identity of each cell.
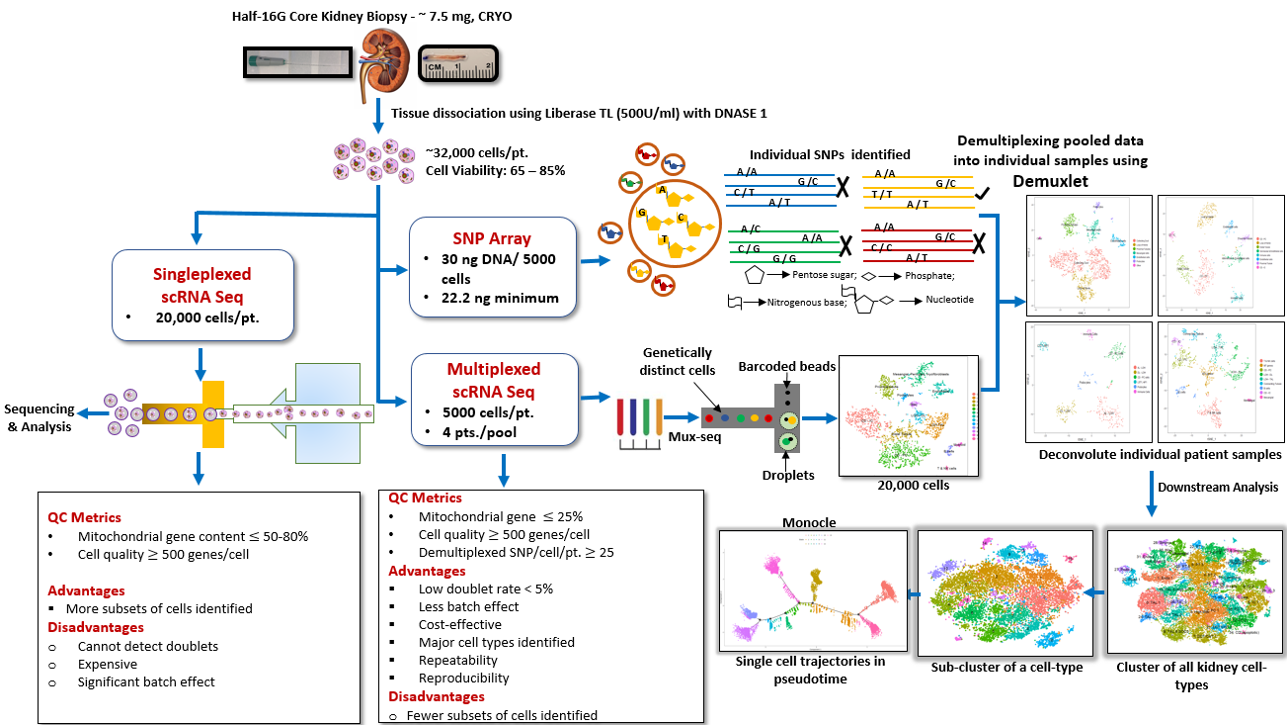
Results: Multiplexed droplet single-cell RNA sequencing results were highly correlated with the singleplexed sample run data. One hundred distinct cell cluster populations in total were identified across the major cell types of the kidney, with varied functional states. Proximal tubular and collecting duct cells were the most heterogeneous, displaying multiple clusters with unique ontologies. Novel proximal tubular cell subsets were identified with regenerative potential. Trajectory analysis demonstrated evolution of cell states between intercalated and principal cells in the collecting duct.
Conclusions: Healthy kidney tissue has been successfully analyzed to detect all known renal cell types, inclusive of resident and infiltrating immune cells in the kidney. Mux-Seq is a unique method that allows for rapid and cost-effective single cell, in depth, transcriptional analysis of human kidney tissue which can be extended to kidney transplant biopsies.
This work was funded by the Kidney Precision Medicine Project (KPMP) Consortium.Juliane Liberto, Parhom Towfighi, Reuben Sarwal and AndréaAlice da Silva assisted in collection of data on the UCSF tissue samples..
