Assessment of HLA typing performance of Korea Biobank Array for transplantation genome-wide association study in Koreans
Sung Min Kim1, Jong Cheol Jeong2, Dong Jin Joo3, Jaeseok Yang4, Myoung Soo Kim3, Hye-Mi Jang1, Hyun-Young Park5, Bong-Jo Kim1, Young Jin Kim1.
1Division of Genome Research, Center for Genome Science, Korea National Institute of Health, Cheongju-si, Korea; 2Department of Internal Medicine, Seoul National University Bundang Hospital, Gyeonggi-do, Korea; 3Department of Surgery, Yonsei University College of Medicine, Seoul, Korea; 4Department of Surgery, Seoul National University Hospital, Seoul, Korea; 5Center for Genome Science, Korea National Institute of Health, Cheongju-si, Korea
Introduction: In clinical transplantation, increased human Leucocyte antigen (HLA) mismatch between donor and recipient were well-known risk factor for graft survival. However, genotyping HLA alleles in a large number of samples is not feasible due to high cost of HLA typing. Alternatively, HLA alleles can be statistically inferred via HLA imputation from Single Nucleotide Polymorphism (SNP) microarray data, the most cost-efficient platform for obtaining genomic data. In 2014, a prospective designed version of the Korean Organ Transplantation Registry (KOTRY) was launched, supported by Korea National Institute of Health (KNIH). For genome-wide association study on liver and kidney transplantation, KNIH is producing more than 7,000 KOTRY samples of genome data using Korea Biobank Array (KBA), a fully customized SNP microarray for Koreans. In KBA, there were more than 10,000 SNPs within HLA regions providing optimal set of SNPs for imputing HLA alleles in Koreans.
Materials and Methods: In this study, we assessed HLA typing accuracy of KBA by comparing HLA imputed alleles from KBA and HLA typing via high-throughput sequencing. Among KOTRY participants, 255 samples were genotyped using KBA and also HLA typed by next-generation sequencing (NGS) using HLAaccuTest kit (NGeneBio, Seoul, Korea). For HLA imputation of KBA, CookHLA was used with a merged Pan-Asian and Korean reference panel of 854 samples. HLA imputation accuracy was assessed by comparing two-digit and four-digit HLA alleles of the imputed and the NGS based typing among HLA-A, HLA-B, and HLA-DRB1 loci, which are the most important three loci for transplantation.
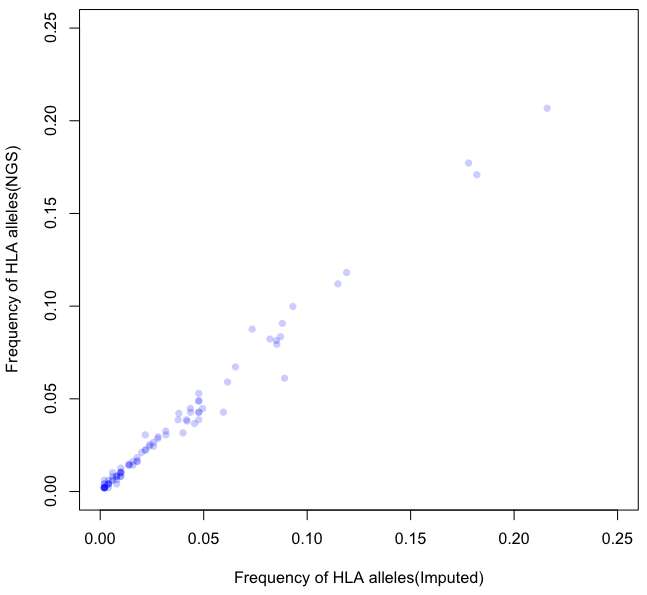
Results and Discussion: In HLA-A, HLA-B, and HLA-DRB1 loci, there were 92 and 93 four-digit HLA alleles for imputation and NGS typing, respectively. Among 84 common HLA alleles, allele frequency was highly correlated (r=0.993) between two typing methods. Next, imputed HLA alleles were compared its true HLA alleles from NGS typing. Overall, accuracy was 97.65% and 92.83% for two-digit and four-digit HLA alleles, respectively. Specifically, accuracy of HLA-A, HLA-B, and HLA-DRB1 two-digit HLA alleles was 99.58%, 95.71%, 97.75%, respectively. For four-digit HLA alleles, accuracy of HLA-A, HLA-B, and HLA-DRB1 was 92.83%, 95.26%, and 89.30%, respectively. Imputed HLA alleles from KBA showed high correlation and high accuracy in an individual level.
Conclusion: Taken together, KBA is an efficient platform to comprehensively obtain 4-digit level imputed HLA alleles with accuracy of about 93%. Although it is not sufficient for clinical practice, imputed HLA alleles would provide sufficient information for large-scale transplantation genome-wide association studies.
This work was supported by an intramural grant from the Korea National Institute of Health (2019-NG-054-00).
[1] Sanghoon Moon and Young Jin Kim et al. The Korea Biobank Array: Design and Identification of Coding Variants Associated with Blood Biochemical Traits. Scientific reports 2019;9:1382
[2] Seungho Cook and Buhm Han. MergeReference: A Tool for Merging Reference Panels for HLA Imputation. Genomics Inform. 2017 Sep; 15(3): 108–111.
[3] CookHLA software: http://software.buhmhan.com/COOKHLA/
There are no comments yet...
