A genome-wide association study identified genetic loci for end-stage renal disease in the Korean population
Hye-Mi Jang1, Hee Jung Jeon2, Sung Min Kim1, Hyun-Young Park3, Bong-Jo Kim1, Jaeseok Yang4, Young Jin Kim1.
1Division of Genome Research, Center for Genome Science, Korea National Institute of Health, Cheongju-si, Korea; 2Department of Internal Medicine, Kangdong Sacred Heart Hospital, Hallym University College of Medicine, Seoul, Korea; 3Center for Genome Science, Korea National Institute of Health, Cheongju-si, Korea; 4Department of Surgery, Seoul National University Hospital, Seoul, Korea
Introduction: End-stage renal disease (ESRD), a kidney failure, is the last stage of chronic kidney disease (CKD). To understand the underlying genetic of ESRD, several genome-wide association studies have been conducted to identify susceptible loci for ESRD. However, a few loci have been discovered yet most of them were hard to replicate in an independent cohort. Moreover, most of previous ESRD GWAS were conducted in samples with European or African ancestry. In 2014, a prospective designed version of the Korean Organ Transplantation Registry (KOTRY) was launched, supported by Korea National Institute of Health (KNIH). For genome-wide association study on liver and kidney transplantation, KNIH is producing more than 7,000 KOTRY samples of genome data using Korea Biobank Array (KBA), a fully customized SNP microarray for Koreans. In this study, we performed a GWAS to identify loci associated with ESRD in the Korean population.
Materials and Methods: All samples of cases and controls were genotyped using KBA with about 830K variants. After quality control, there were 1,840 samples of ESRD cases from the KOTRY and 54,370 controls from the Korean Genome and Epidemiology Study (KoGES). Controls were selected as non-ESRD samples without type 2 diabetes and hypertension. After quality control on variants, variants with minor allele frequency >= 1% were selected and imputed using IMPUTE v4 with the merged reference panel of 1,000 Genomes project (n=2,504) and Korean Reference Genome (n=397). Logistic regression was performed by adjusting age and gender using EPACTS software.
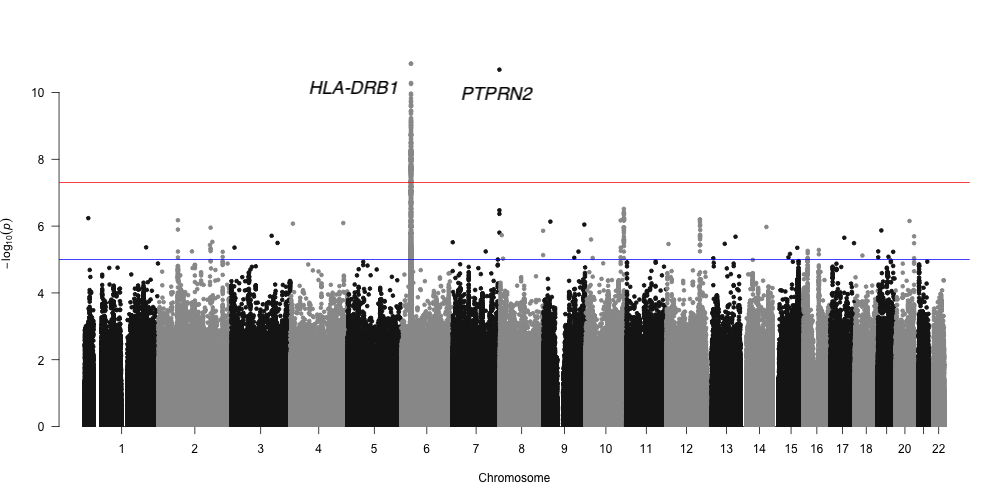
Results and Discussion: At the genome-wide significance (P < 5x10-8), two loci including HLA-DRB1 and PTPRN2 were associated with ESRD. The locus HLA-DRB1 was previously associated with estimated glomerular filtration rate (EGFR) in Europeans. The locus PTPRN2 was reported to be associated with serum metabolite concentrations in CKD in Europeans. However, none of previously reported ESRD loci was replicated in this study at the level of suggestive statistical significance (P < 1x10-6).
Conclusion: In this study, we performed GWAS on ESRD in Koreans and identified two ESRD candidate loci including HLA-DRB1 and PTPRN2. These loci were previously reported to be associated with EGFR and serum metabolite concentrations in CKD in Europeans. However, replication study in an independent cohort is warranted to validate our findings.
H.M.J. and H.J.J. contributed equally to this work. J.Y. and Y.J.K. are corresponding authors.. This work was supported by an intramural grant from the Korea National Institute of Health (2019-NG-054-00)..
[1] Maria P. Hernandez-Fuentes et al. Long- and short-term outcomes in renal allografts with deceased donors: A large recipient and donor genome-wide association study. Am J Transplantation 2018;18:1370–1379.
[2] Sanghoon Moon and Young Jin Kim et al. The Korea Biobank Array: Design and Identification of Coding Variants Associated with Blood Biochemical Traits. Scientific reports 2019;9:1382
There are no comments yet...
