
Can epitope matching outmatch traditional HLA matching?
Christian Unterrainer1, Matthias Niemann2, Nils Lachmann3, Caner Süsal1.
1Institute of Immunology, University of Heidelberg, Heidelberg, Germany; 2PIRCHE , AG, Berlin, Germany; 3Center for Tumor Medicine, Charité University Hospital, Berlin, Germany
Collaborative Transplant Study.
Introduction: Pre-transplant presence and post-transplant development of alloantibodies against highly polymorphic human leukocyte antigen (HLA) allele mismatches (mm) play a major role in rejection of kidney allografts. However, antibodies recognize small sequences on HLA alleles called epitopes and the same epitope can be present on different alleles, impairing the precision of matching. Using the Collaborative Transplant Study database, we analyzed whether a T-cell epitope-based matching of donor-recipient pairs using the PIRCHE-II algorithm is a better predictor of graft survival than the currently applied allele group-based HLA-A, -B, -DR matching.
Methods: 68,606 European deceased donor kidney-only transplantations during 1996–2016 were analyzed. PIRCHE-II mismatch score was calculated based on split typing using the 2007 NMDP haplotype frequency database and the PIRCHE matching service. Multivariable Cox regression analyses were performed to calculate hazard ratios (HRs) for death-censored graft loss. Correlation was analyzed with Spearman’s rank correlation. In order to achieve comparable HRs, a log-transformed PIRCHE-II score was used, followed by a linear transformation to match the value range of 0–6 HLA A+B+DR mm. Linear transformations in Cox regression analysis change the HR per point without changing the P- and Z-values.
Results and Discussion: The HLA mismatch-derived PIRCHE-II score correlated strongly with the number of HLA mm (correlation coefficient ρ=0.65, P<0.001). Cox regression analyses revealed that Pirche II score and HLA mm are predictive confounders for death-censored graft loss at year 5 with highly significant prognostic power (Z-value for Pirche II: 9.8; for HLA: 11.2; P<0.001) (Table 1).
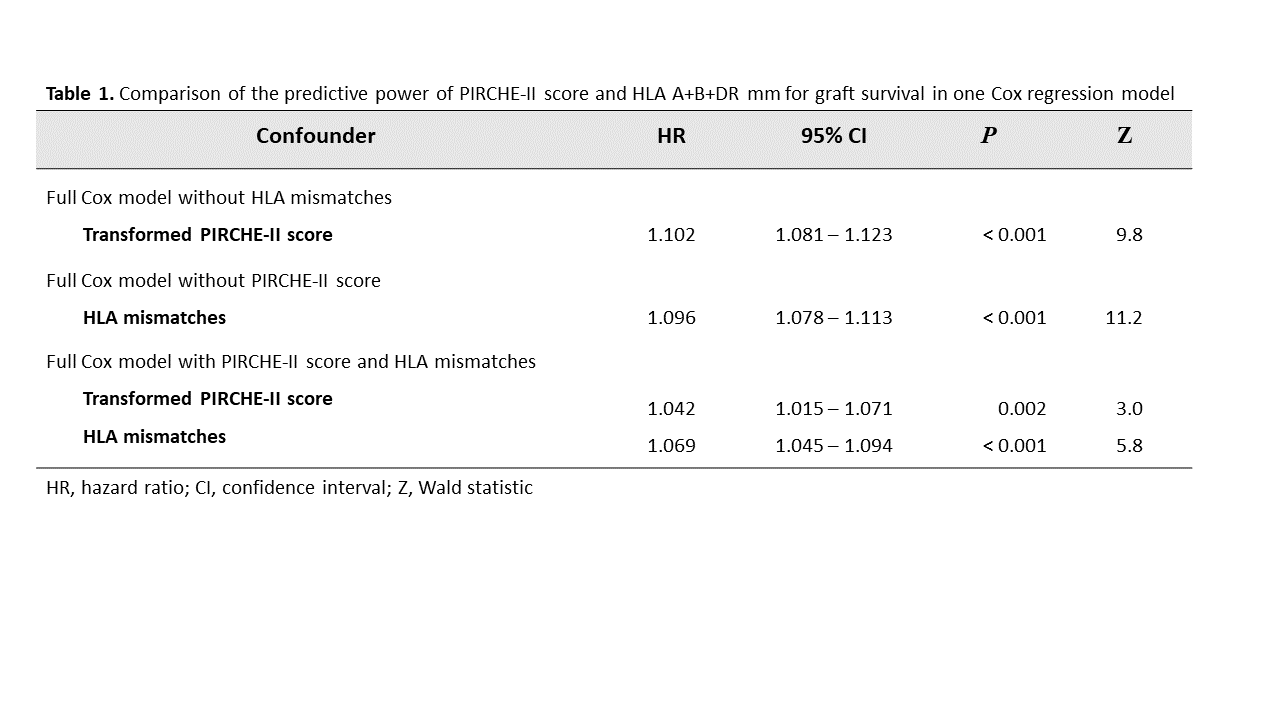
The full Cox model per transformed PIRCHE-II score without HLA mm resulted in a hazard ratio (HR) for graft loss of 1.102 (95% CI=1.081–1.123; P<0.001). When HLA mm were added to the model, the HR per PIRCHE-II score decreased to 1.042 but remained statistically significant (95% CI=1.015–1.071; P=0.002) (Table 1). Overall, considering the HLA loci currently used for typing at split antigen level, HLA mm still had a higher predictive value than the PIRCHE-II score (11.2 vs. 9.8) regarding graft survival. As illustrated in Figure 1, the variance in PIRCHE-II score increased with higher number of HLA mm.
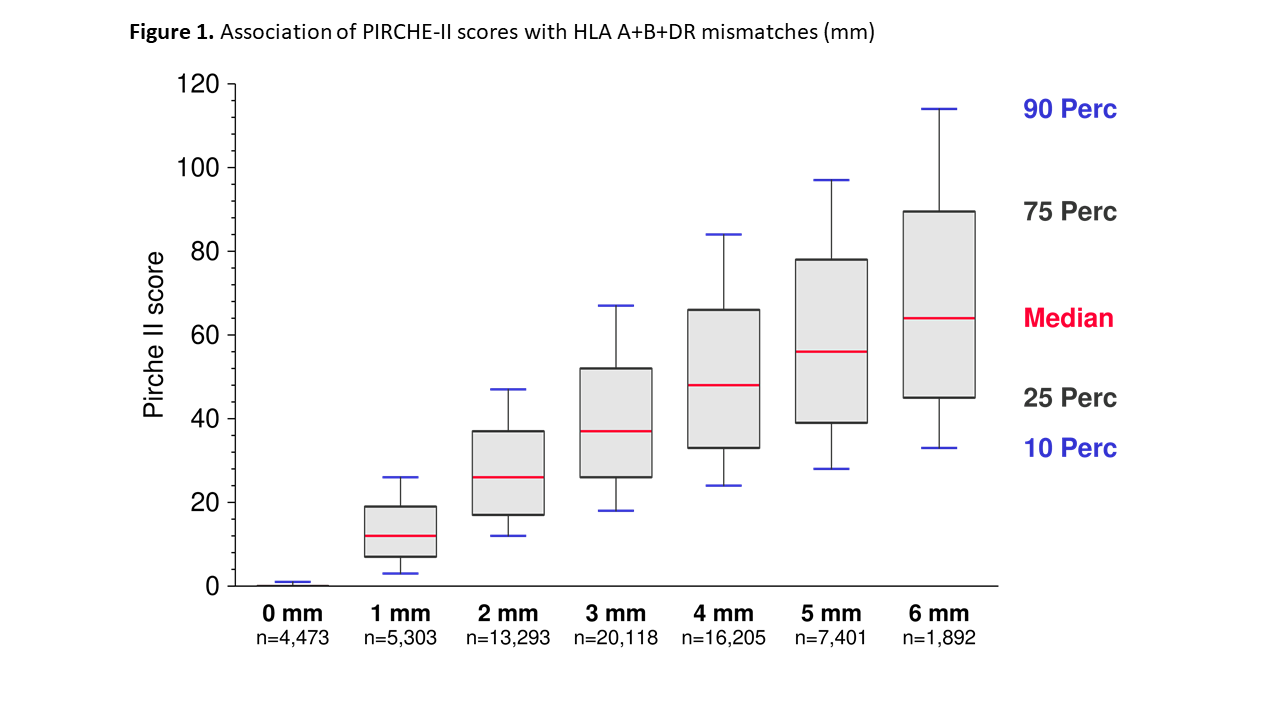
Therefore, the possible clinical benefit of PIRCHE-II score matching improved with higher number of HLA mismatches. As demonstrated in Table 1, the risk of graft loss increases with each additional PIRCHE-II score.
Conclusion: Our results indicate that PIRCHE-II score is useful and can be included into matching algorithms. However, it cannot fully replace traditional HLA matching. There seems to be information on immunological differences PIRCHE-II score cannot cover in its present form using HLA typing data at split level. This needs to be analyzed also for HLA matching at 4-digit molecular typing level.
There are no comments yet...